Ontology-Guided Machine Learning
Project Description | Background | Team Members | Acknowledgement | Publications | Research
Research Project Introduction
Personalized evidence-based medicine, along with the unprecedented growth in volume and complexity of biomedical data (the availability of Big Data), calls for the use of new intelligent technologies. While many computational scientists and researchers in areas such as Machine Learning (ML) or Data Mining (DM) focus on the ability to procesismbs massive amounts of data and build accurate models, the complexity, heterogeneity and semantics of biomedical data are often outside of the mainstream research. Healthcare is particularly rich in domain knowledge and that knowledge has been formally represented by using ontologies such as the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT), International Classification of Diseases (ICD), and Unified Medical Language System (UMLS).
This research aims at advancing computational and machine learning methods in order to achieve better applicability in real-world biomedical applications. Specifically, the goal is to develop an ontology-guided ML method to promote the effectiveness of data analytics in healthcare. The ontology-guided ML program involves the use of ontology and verifiable inferences based on the ontology to effectively analyze the complex and heterogeneous biomedical data. The method will be applied to large and complex dataset called SEER-MEDICARE and SEER-MHOS.
Project Description
The project aims to create and test predictive Machine Learning (ML) methods capable of computing rates of mortality and comorbidities, as well as quality of life for cancer patients. Two separate datasets will be used in order to compute these two models. The first dataset SEER-MEDICARE is based on prostate cancer diagnosis which will be used to compute mortality and comorbidity rates. The second dataset SEER-MHOS is based on prostate, breast, colorectal, lung and bronchus, uterus, bladder, head and neck, melanomas – skin, stomach and pancreas cancer diagnosis which will be used to predict patient quality of life and comorbidities.
Background
AQ21 | SEER-MEDICARE | SEER-MHOS
AQ21
AQ21 is a Machine Learning (ML) program that is based on Natural Induction. For this research project, we will use AQ21 to generate predictive healthcare models. To incorporate healthcare data, Unified Medical Language System (UMLS) domains will be added to AQ`s knowledge base. This integration is referred to as an ontology-guided AQ21, once its domains are set we will begin creating predictive models using patient datasets (SEER-MHOS and SEER-MEDICARE). For more information on ML program AQ click here
SEER-MEDICARE
The SEER-MEDICARE data contains prostate cancer patient information. The research study will predict mortality and comorbidity rates based from the initial diagnosis to any moment after the discovery of the prostate cancer occurrence. The datasets will be linked to hierarchies in the Unified Medical Language System (UMLS). This will allow us to compute the relationships and process the data with the ontology-guided AQ21 program.
The figure below presents the SEER-MEDICARE dataset timeline:
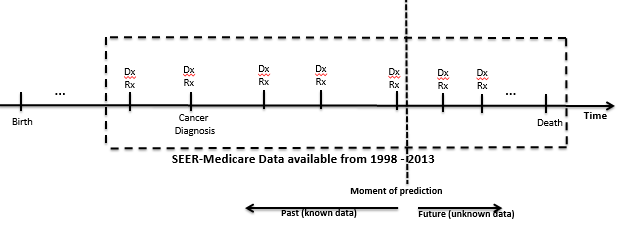
Hypothesis: Comorbidity, demographics, treatment type, complications, medications, and cancer specific data can accurately predict mortality and future comorbidities and complications. Moreover the ontology-based machine learning method will achieve higher accuracy than non-ontology based methods.
SEER-MHOS
SEER-MHOS is a semi-structured dataset, that contains patient cancer information. This research will specifically focus on following cancer sites: prostate, breast colorectal lung and bronchus, uterus, bladder, head and neck, melanomas, stomach and pancreas. SEER-MHOS data will be transfrered into a structured dataset using a universal ontology, such as UMLS. Once this stage is completed the ontology-guided AQ21 program will analyze the data and create predictive models. For this particular research our aim will be at predicting Activities of Daily Living(ADLs), Physical Component Summary(PCS) and Mental Component Summary(MCS), and Comorbidities following cancer diagnosis.
Hypothesis: Comorbidity, demographics, and cancer specific data can accurately predict ADLs, Physical Component Summary (PCS) and Mental Component Summary (MCS) and future comorbidities.
Team Members
 |
 |
| Dr. Hua Min, Assistant Professor of Department of Health Administration and Policy, George Mason University. |
Dr. Janusz Wojtusiak, Associate Professor of Health Informatics and Director of Machine Learning and Inference Laboratory, George Mason University. |
 |
 |
| Katherine Irvin, MLI Research Assistant, PhD student in Information Technologies, George Mason University |
Hedyeh Mobahi, Research Assistant and Graduate student in Health Informatics, George Mason University. |
 |
 |
| Sava Vukomanovic, Research Assistant and Undergraduate student in Applied Information Technology, George Mason University. |
Ilirjeta Krasniqi, Research Assistant and Undergraduate student in Health Administration and Policy, George Mason University. |
-Applying an Ontology-guided Machine Learning Methodology to SEER-MHOS Dataset, Bio-Ontologies SIG
-Applying Machine Learning Methods to Predict Activities of Daily Living for Cancer Patients, AMIA
–Visualizing Effects of Cancer on Relationships Between Comorbidities and Activities of Daily Living, AMIA
-Visualizing the Effects of Cancer on Patients overall Quality of Life, Georgetown University URC
Peer-reviewed Papers in Conference and Workshop Proceedings
Min H., Mobahi H., Vukomanovic S., Irvin K., Krasniqi I., Avramovic S., and Wojtusiak J., “Applying an Ontology-guided Machine Learning Methodology to SEER-MHOS Dataset,”, 2016 Bio-ontology at Intelligent Systems for Molecular Biology(ISMB), Orlando, Florida, July 8-9, 2016.
Wojtusiak J., Min H., Elashkar E., Mobahi H., Vukomanovic S., “Ontologies in Supervised Learning from Medical Data”, 4thArtificial Intelligence for Knowledge Management (AI4KM), July 9, New York City, 2016 (Invited paper)
Peer-reviewed Abstracts in Conference and Workshop Proceedings
Min H., Oz T., Vukomanovic S., Mobahi H., Irvin K., Krasniqi I., Wojtusiak J., “Visualizing the Effects of Cancers on Relationships Between Comorbidities and Activities of Daily Living”, 2016 AMIA Annual Symposium, November 12-16, 2016, Chicago, IL, Accepted.
Min H., Oz T., Vukomanovic S., Mobahi H., Irvin K., Krasniqi I., Wojtusiak J., “Applying Machine Learning Methods to Predict Activities of Daily Living for Cancer Patients”, 2016 AMIA Annual Symposium, November 12-16, 2016, Chicago, IL, Accepted.
Min H., Mobahi H., Vukomanovic S., Irvin K., Krasniqi I., Avramovic S., Wojtusiak J.,“Ontology applications in Machine Learning”, 2016 Bio-ontology at Intelligent Systems for Molecular Biology (ISMB), Orlando, Florida, July 8-9, 2016.
Student Poster
Vukomanovic S., Krasniqi I., Mobahi H., Min H., Wojtusiak J., “Visualizing the Effects of Cancer on Patients Quality of Life”,Georgetown University Undergraduate Research Conference, Department of Human Science at the School of Nursing & Health Studies, April 14-15, 2016
Acknowledgement
Funding for this project is provided by the Jeffress Trust Awards Program in Interdisciplinary Research.
References
Min H., & Wojtusiak, J., “Clinical data analysis using ontology-guided rule learning.” In Proceedings of the 2nd international workshop on Managing interoperability and complexity in health systems, pp. 17-22. ACM, 2012
Wojtusiak, J., “Semantic Data Types in Machine Learning from Healthcare Data,” Proceedings of the International Conference on Machine Learning and Applications (ICMLA), Florida, December, 2012
Wojtusiak, J., Michalski, R. S., Kaufman, K. & Pietryzkovski, J., “The AQ21 Natural Induction Program for Pattern Discovery: Initial Version and its Novel Features,” Proceedings of The 18th IEEE International Conference on Tools with Artificial Intelligence, Washington D.C, November 13-15, 2006.
Wojtusiak, J., “AQ21 User`s Guide,” Reports of the Machine Learning and Inference Laboratory, MLI 04-3 George Mason University, Fairfax, VA, September, 2004
Additional references are available in the publications section.
